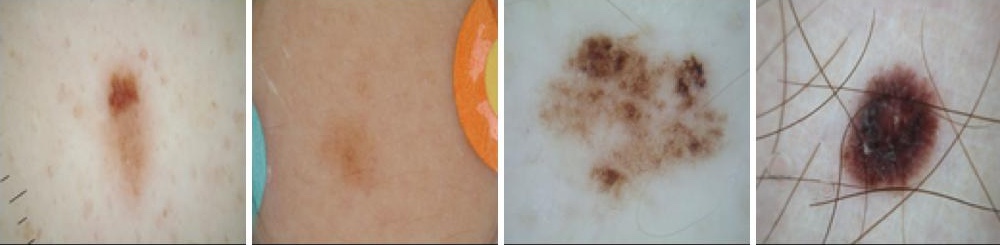
This project focuses on analyzing skin lesions using the ISIC 2018 dataset. I am using U-Net for lesion segmentation and ResNet50 for classifying lesions into different types.

This project focuses on analyzing skin lesions using the ISIC 2018 dataset. I am using U-Net for lesion segmentation and ResNet50 for classifying lesions into different types.
This project predicts heart attack risk using clinical data and machine learning, focusing on implementing and exploring Snowpark in Snowflake for data processing, model training, deployment and inference.

I developed an app for processing user-provided text using Transformers. It uses BART for summarization, Pegasus for paraphrasing, DistilBERT for named entity recognition, KeyBERT for keyword extraction, and WordNet for finding synonyms.
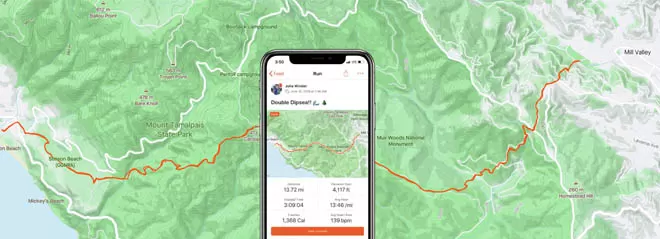
I created an automated ETL pipeline using Python to extract data from the Strava API, transform it, and load it into a PostgreSQL database on a weekly schedule. The process is automated with Airflow, which runs in Docker containers, to ensure regular and efficient updates.
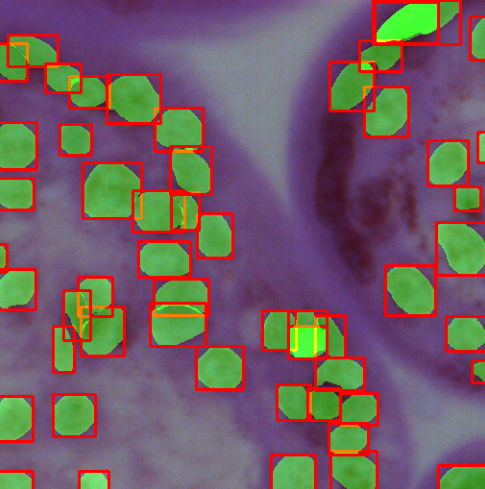
Developed an end-to-end cell segmentation system using the YOLO v8 architecture to accurately segment and classify white and red blood cells from microscopic images. The model was integrated into a web app created using Flask and deployed on Azure.

Developed a recipe recommendation system that uses NLP and web scraping to suggest recipes based on provided ingredients. It matches ingredients with recipes from multiple sources to simplify meal planning and reduce food waste.
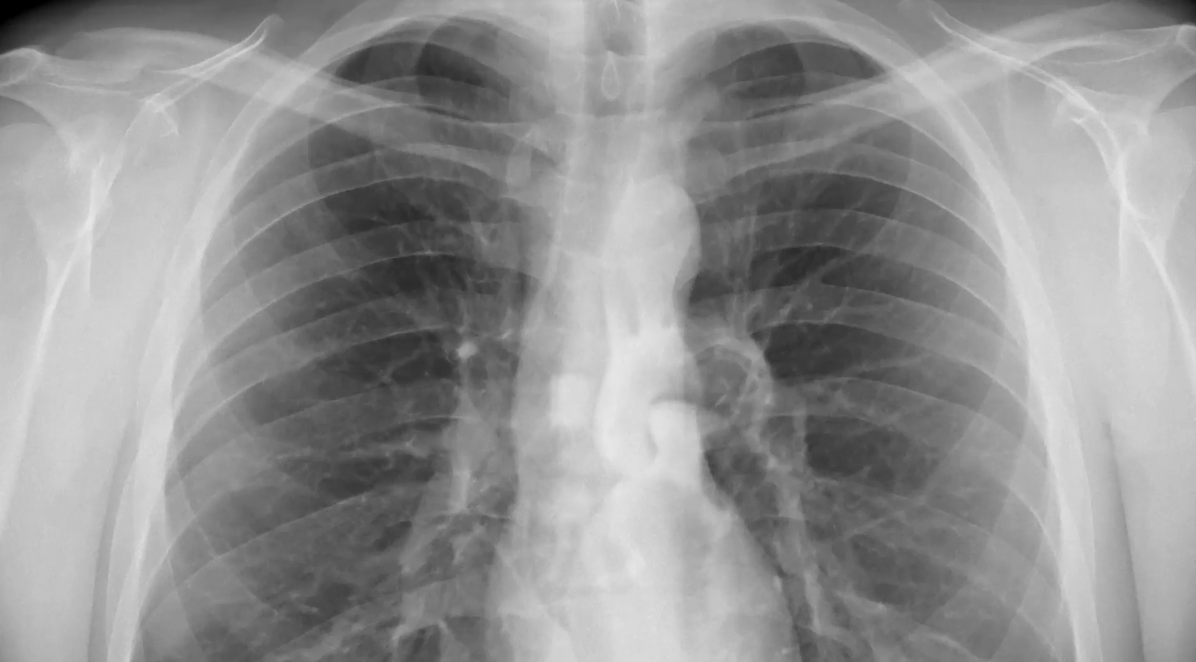
Developed neural network models to detect pneumonia from chest X-ray images using CNN and InceptionV3 architectures. The models achieved F1-scores of around 90% and 95% on the test set, respectively.

I developed a classifier to identify the actors from the "Friends" TV show in images. Using OpenCV, I preprocessed the images to extract facial features. I then trained a machine learning algorithm to classify the faces based on these features.
Developed an algorithm that can predict the course of multiple sclerosis in a specific time window, considering results from a single visit and sequences of consecutive visits (time series). This was achieved by using classical ML methods and a LSTM NN. The models achieved F1-scores ranging from 71% to 77%. Several explainability methods were also applied.
Project developed to predict the outcome of Covid-19 disease in South Korea using patient data combined with background data, such as weather, region and policy decisions. The real data was cleaned and preprocessed, including scaling, feature reduction (PCA & LDA) and feature selection. Multiple classifiers, including SVM, KNN and Naive Bayes, were tested, achieving a F1-Score of 96%.
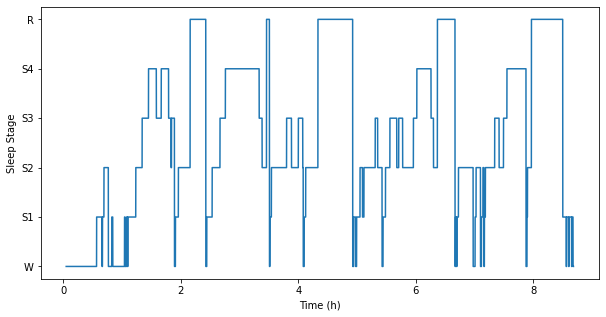
Analyzed and processed different physiological signals (EEG, ECG, EMG and EOG) acquired during a whole night of sleep. Used techniques such as ICA, Filtering, Wavelets, PCA, and MDS. Developed an unsupervised clustering method to identify and classify the different sleep stages.
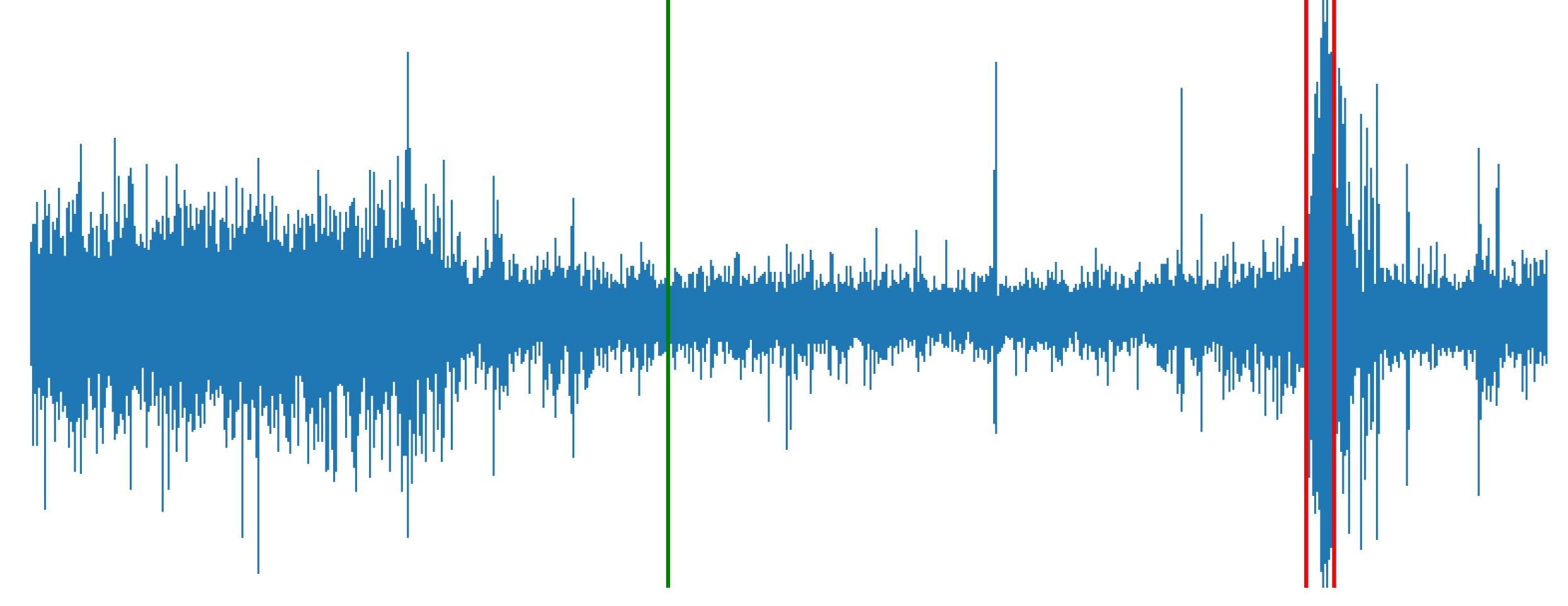
This study aims to predict and detect seizures using EEG data. I focused on identifying patterns within the three classes: Interictal, Pre-Ictal, and Ictal, using neural networks (CNN and LSTM).
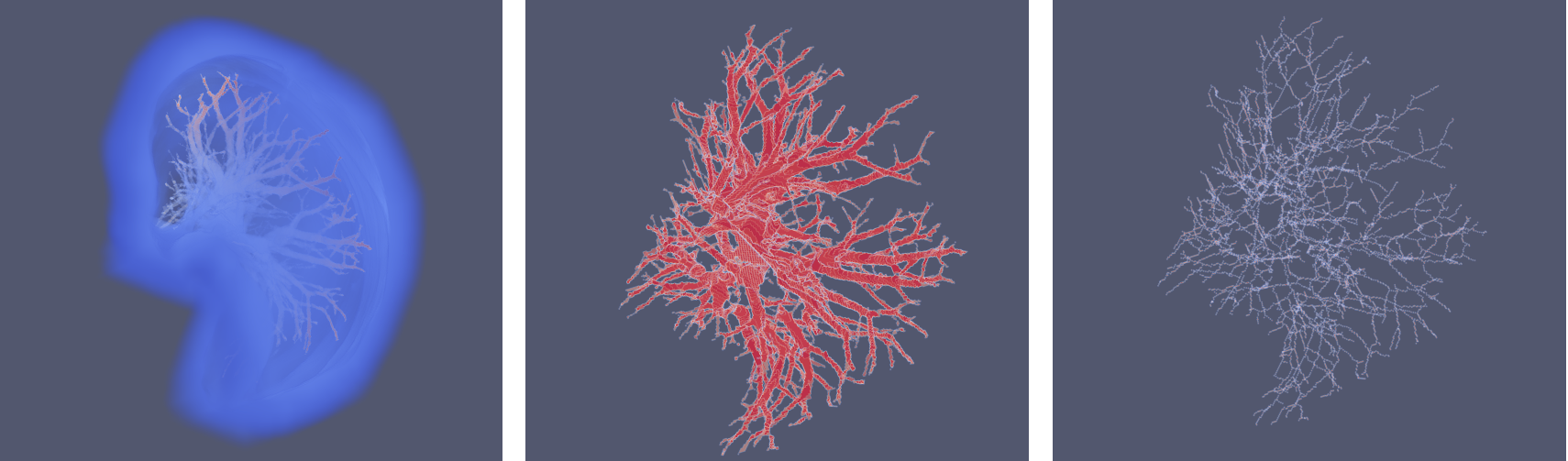
Project developed to calculate the number of vessels, their diameter, and the number of bifurcations from 3D files of a vascular network.
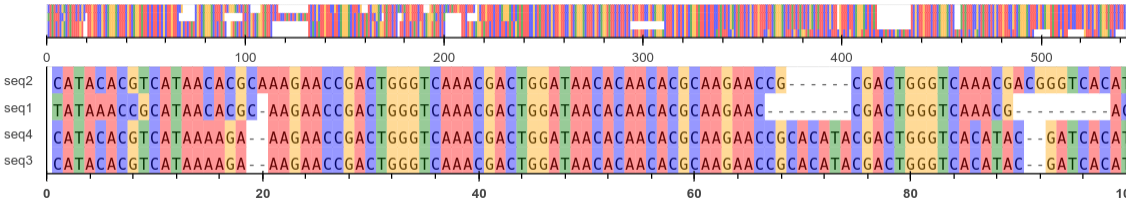
A variety of bioinformatics algorithms were developed as part of an exercise, addressing tasks such as graph analysis, sequence alignment, gene expression analysis, and protein structural predictions.